PCR Biological Method Overview
PCR, or polymerase chain reaction, is a method to amplify a segment of DNA for analysis. Because it is such a powerful technique, there are a HUGE number of situations where PCR may be used. Some common reasons for using it are:
- Microbiology: You need to know if your bacteria was transformed properly with your plasmid
- Oncology: You need to know if a particular gene exists in your cancerous cells
- Forensics: You need to know if a certain criminal’s DNA was present at the crime scene
The basic steps of PCR include:
- Designing primers to designate a target DNA sequence to amplify
- Mixing together the primers with the target DNA strand, polymerase enzyme, and deoxynucleotides
- Running a thermocycler multiple times in “cycles” to repeatedly:
- Separate DNA strands
- Allow primers to anneal
- Allow the polymerase to attach and synthesize a new DNA chain
- Sequencing or Electrophoresis to prove that the PCR worked
In a nutshell, here is what Step 3, above, looks like:
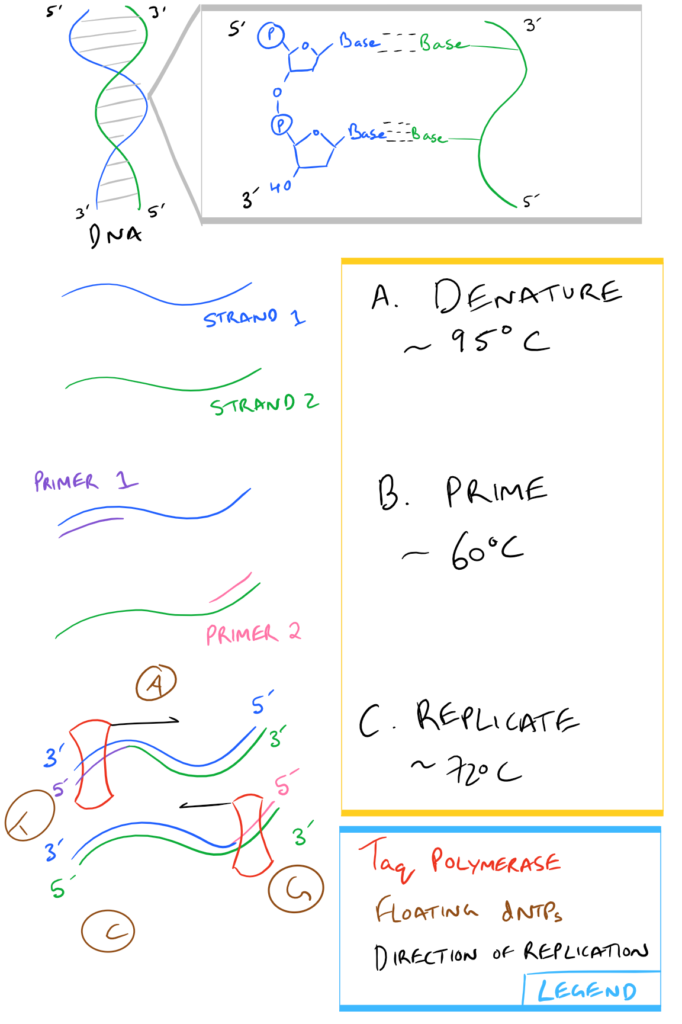
Note: If you’re writing research papers, I highly recommend Grammarly – it’s a free grammar check plugin for Chrome. Try it out here…
Ingredients in a PCR mixture – What every component does
In general, a PCR tube will contain the following items:
| Water: | Solvent |
| dNTPs (or deoxynucleotide triphosphates): | Single bases A, T, C, and G which are used by the polymerase while replicating the DNA. As the polymerase adds base pairs onto the new DNA strand, one base pair is used at a time. |
| MgCl2: | An essential cofactor for the polymerase enzyme |
| Primers: | Short segments of single-stranded DNA used to frame the DNA region that needs to be amplified. They are complementary to the template DNA strand only at defined locations around the target sequence. |
| Target DNA: | The DNA “template” that you want to make copies of. This can be a full DNA chain or a part of a longer chain. |
| Taq Polymerase: | An enzyme from Thermis aquaticus that uses dNTPs and replicates DNA starting from the 3′ end of a template strand towards the 5′ end. Taq is used in PCR specifically because it is resistant to the high temperatures used for separating DNA strands during Step 3a above . |
Complete Step-By-Step PCR Protocol
Materials
|
PCR tubes |
|
|
PCR temperature Cycler |
|
|
Pipettes |
|
|
Reaction Buffer |
10 mM Tris-HCl, 50 mM KCl, and 1.5 mM MgCl2, pH 8.3 |
Note: Grammarly is a free grammar check plugin for Chrome. I used it for this article and really like it! Try it out here…
Methods
- For each PCR tube set up the following mixture of materials up to a 50 ul total volume on ice
dNTP solution
200 uM for each dNTP
Forward Primer
0.2 uM
Reverse Primer
0.2 uM
Target DNA
Less than 1000 ng
Taq Polymerase
1.25 units per tube
Nuclease Free Water
Q.s up to 50 ul
Mineral Oil
Add a little bit on top of each tube if you don’t have a heated lid on the temperature cycler
- Pipette each tube up and down several times, gently, so as not to add bubbles
- Centrifuge each tube down for 1-2 seconds at 100 rcf to bring all contents to the bottom
- Heat up the temperature cycler to 95oC
- Quickly transfer tubes from ice to the temperature cycler and begin thermocycling
- Typical thermocycle procedure:
Step Name
Temperature
Time
Denaturation
95 oC
30 seconds
Cycle Denaturation
95 oC
30 seconds
Cycle Priming
50-60 oC
60 seconds
Cycle Extension
72 oC
1 minute per kilobase of target DNA
Final Extension
72 oC
5 minutes
Hold Temperature
4 oC
Infinite
- Repeat the above “cycle” steps 35-45 times.
- Make sure to keep the samples on ice while you are not using them. The infinite hold sequence allows you the flexibility of leaving your samples in the temperature cycler while you are busy with other lab work.
Notes on this PCR Methodology
- Note 1. It is important to design your PCR primers to be specific to only the regions flanking the target sequence. Typically, specific primers are ~30-40 bases in length.
- Note 2. The Tm (melting temperature) of the primers affect the temperature in Step 3b and the “Cycle priming” step.
- Note 3. For greater accuracy/fidelity while copying DNA, a Pfu polymerase (from Pyrococcus Furiosus) may also be used.
- Note 4. With 35 cycles, the target DNA is amplified 236 times its initial concentration. For times when you have very little target DNA, you can amplify 45 times or more.
- Note 5. For Primer design, you can use tools such as Primer 3. Tools are also available for calculating the Tm of your primers such as this Tm calculator.
- Note 6. G-C rich sequences will need longer denaturation times (typically up to 5 minutes) because of the additional hydrogen bonding. Any template with more than 60% G-C bases is considered G-C rich.
[…] and isolation is a precursor for many methods in molecular biology including Northern Blotting, RT-PCR, and Microarray analysis. This blog post will focus on this precursor method as opposed to other […]
[…] your sticky ends and it will form a phosphodiester linkage. If this is confusing, check out the Polymerase chain reaction (PCR) guide for images on what DNA looks like. This is shown […]